(Big) Data Journalism with Spark and R
This post shows how to process gigabytes of unstructured data with the Spark framework and using the R language.
At Swiss Public TV and Radio (SRF) we recently published an investigation of the “Collection #1-5” password leaks. We showed that more than 3 million Swiss email addresses and – more disquietingly – over 20’000 email addresses of Swiss authorities and providers of critical infrastructure appear in the leak (text in German).
So basically, the problem boiled down to preprocessing, filtering and searching through a very, very large data set. Some journalists prefer to cooperate with external experts when it comes to such problems. This is understandable, as many lack the time and knowledge to do it on their own. However, I think that tackling such problems wholly inside newsrooms has one tremendous benefit: You completely control what is done with the data, and you know exactly what can be interpreted into it – and what not.
In this blog post, I don’t want to get into too much code-specific detail, as I don’t want people misusing this research for malicious purposes. Also, I won’t talk about the context of the password leaks and such. Wired has a good article that summarizes it well.
In lieu, I’d like to describe some parts of my approach: First of all because it’s part of our DNA at SRF to publish very detailed methodological descriptions of what we do.
Secondly, because I think dealing with large, complex data sets becomes more and more daily business in data journalism. Thus, I hope other (data) journalists can benefit from my learnings and my advice.
Here’s the structure of this admittedly quite long blog post:
- Was I actually dealing with big data or am I just bragging?
- The hardware and software setting I used
- Intro to Spark
- How R can communicate with Spark
- The logic behind Spark “jobs”
- “How I searched for the Needle in the Haystack”: My pipeline in three major steps from preprocessing to analysis
- Summary and some advice
“Big” Data? Srsly?
Well, you know. We’ve all had our fair share of laughs when people would come to us and think of us data journalists as the guys dealing with “big data”, when our most data-ambitious projects weren’t going over 200MB or so. There’s still a big misunderstanding about that term, I think. Anyway, let’s quickly do the 5-V-check on the “Collection #1-5”:
- Volume. Well, 900GB+ certainly don’t fit into memory. And they don’t fit on my laptop’s SSD drive, either. Not even on the SSD of our workstation (see below). I had to buy an HDD. So that’s a starter.
- Velocity. Okay, zero velocity here. I downloaded the data, and then it stayed like it was when I had downloaded it. That’s a negative.
- Variety. A point for big data here. The password leak has tens of thousands of single files, most of them .txt, but also many others, like .sql, .csv, Archives, etc. Also, while the structure of the files is mostly similar (email, delimiter, password, one per line), there’s still some annoying variety in there.
- Veracity. The veracity of the different leaks inside the collections can not be taken for granted – quality and accuracy are certainly varying, there are many duplicates, for example. Troy Hunt has a nice blog post on that. too. So, another point for big data.
- Value. Let’s leave that to the black-hat hackers and security experts, shall we?
So I can – with good faith – make a point that I was dealing with something that could be coined “big data”. I know some people will disagree, that’s why I put the “big” in the title in parentheses.
The setting
I’ve already talked about the data at hand. For the whole process, I used a workstation with the following specs:
- 2 CPUs with 10 cores each (results in a total of 40 threads)
- 64GB RAM
- 512GB SSD (of which about 280GB were free)
- 6TB HDD
- Linux Ubuntu 18.04 with Spark 2.4.x and R 3.5.x
For various reasons, among which is the sensitivity of the data, I decided against using a cloud-based service such as Azure, Google Cloud or the like.
What was clear from the beginning was that I couldn’t use my ordinary R data processing pipeline, for the simple reason that the data would never fit into memory, and I would have to use some tedious batch processing logic that would only process parts of the data at once (which I ended up doing anyway, but only for parts of the process). Also, I finally wanted to try out Spark in “production”.
Spark?!
So Spark is one of those “big data technologies” that gained a lot of traction in the last few years. Being developed by the Apache software foundation, it is “an open-source cluster-computing framework […] with implicit data parallelism.” So, in simple terms, Spark is about parallelizing data processing tasks over a cluster, i.e. a network of computers that each processes a task (preferably in-memory, so it’s quite fast). In my case, the “cluster” was the workstation, and the cores (or threads) were the “network of computers”.
Probably one of the most important things about working with Spark is to keep disk input/output low (i.e. from disk to memory and back). And of course, it only works with tasks that can be parallelized. Applying the same preprocessing steps to billions of lines in text files is such a task.
To me, as a layman, the most obvious advantage of using Spark was having not to do deal with out-of-memory (OOM) errors all the time. In the second major step of the project, I had to start jobs with ~300GB+ (all at once, no batch processing) and could leave them running without having to worry about OOM or similar. Spark has a nice web UI that shows the progress of running jobs and gives a lot of detail about single stages and tasks.
I’ve been using the terms “jobs”, “stages” and “tasks” above. In Spark, “job” is the general term for a set of data transformations that result in a so-called “action”, i.e. writing to disk or sending the (condensed) results of a query to a frontend like R. A job is broken up into “stages“, each roughly corresponding to a set of transformations that can be run without “shuffling” data between nodes. If such shuffling is required, a new stage begins. In turn, each stage is broken up into “tasks“. For example, if 100MB need to be processed in a job, and there are 40 nodes, like in my case, ideally, the 100MB are split up into 2.5MB each. So there are 40 tasks that have to be processed by the cluster, one task per node (with 40 nodes). After each node has finished its task, the job is more or less done. If the data would be 200MB big, one could have either 80 tasks (i.e. two tasks per node that are processed after each other) or increase the task size.
You see that there are quite some parameters involved, e.g. the minimum task size or how stages are split up into tasks. For laymen like me, this can be delegated to Spark, and in many cases, it will work fine.
R and sparklyr
Prominent languages for interacting with Spark are Java and Scala, probably followed by Python (through PySpark). Recently, support for R has grown, mostly through the development of the sparklyr package which is supported by RStudio. There is also SparkR, which might be just as good.
I chose to work with sparklyr, mainly because it allows to use the well-known dplyr declarative syntax to interact with Spark. Behind the scenes, the dplyr queries are translated to SQL queries that are executed on the Spark cluster
One of the most important concepts to understand is that sparklyr queries evaluate lazily. In the words of the official documentation, it “never pulls data into R unless you explicitly ask for it” and “it delays doing any work until the last possible moment: it collects together everything you want to do and then sends it to the database in one step”. That’s a major advantage of working with Spark.
A concrete example of a Spark job
Let’s go through a whole job from the third major step of my pipeline (see below), in order to understand this concept:
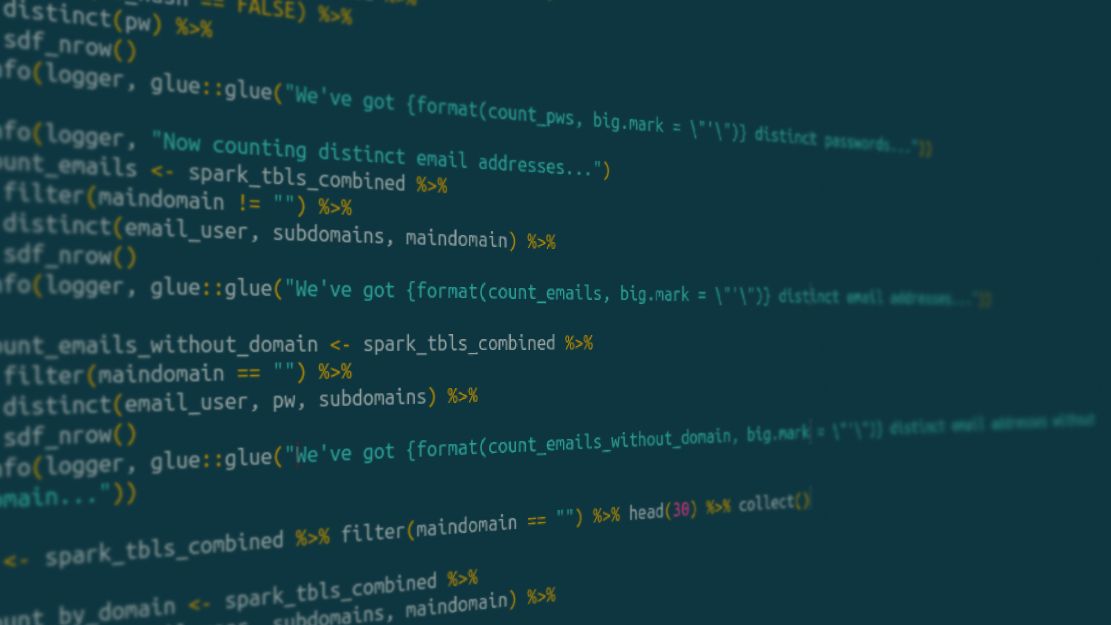
spark_tbls_combined %>%
filter(maindomain != "") %>%
distinct(email_user, subdomains, maindomain) %>%
sdf_nrow()
In this simple dpylr/sparklyr pipeline, I first want to remove entries in my data frame where the field “maindomain” is empty (filter()). I then want to count the distinct combinations of “email_user”, “subdomains” and “maindomain”. For this, I use the distinct() function followed by the sdf_nrow() function (which is sparklyr-specific and worked better for me than the well-known count function). “spark_tbls_combined” is a variable that basically holds a reference to four files that were loaded into memory before, totaling about a gigabyte of data.
Spark needs three stages for that job, as can be seen from the Spark Web UI:

It can also be seen that the first stage (very bottom) is split up into 714 tasks, and “reduces” the data size to 216.3MB, which is then split up into 2001 tasks in the second stage, and so on. You can also look at the execution plan for the SQL query that is executed on the data:

A short note on performance: We’re talking about 31 million lines of data here (all lines with Swiss email addresses in the leak). On our workstation, reducing this to the distinct email addresses (approx. 3,3 million) took a mere 10.099 seconds (4 + 6 + .099). Of course this is due to parallelization and so forth, but what’s also important to understand is that Spark has a very good query optimizer. The query above is very simple, but if you have a more complicated, more costly query, Spark will try to optimize it and build the most efficient physical execution plan as possible. For example, Spark will try to do “predicate pushdown”, i.e. try to filter the data as early as possible to reduce data size (even though your filter function may come very late in the dplyr pipeline).
So not until all the query processing in Spark has taken place, the data is returned to R. In this case, only a single number is loaded into R (the count of distinct addresses). This is very important: To process as much data as possible in Spark and load only what’s necessary into R, because R can only handle as much data as there is memory – and of course doesn’t have any parallel processing capabilities (natively). You’re free to further process Spark results in R, taking full advantage of the more familiar R commands you know. If it fits well into memory and/or has a mere several thousand records, that shouldn’t be a problem.
How I searched for the Needle in the Haystack: Preprocessing
In this section, I’d like to give a high-level overview of how I reduced a huge collection of ten thousands of different files with different structure to a tidy Spark data frame. This data frame had several columns such as “maindomain” and “subdomains” so I could easily do group-by-operations later, for example, in order to find out that more than 500 email addresses in the leak belong to “vtg.admin.ch”. Which is the email domain of the Swiss Armed Forces. In simple steps, this preprocessing involved:
- Sanitizing file names: There were a lot of non-latin characters in the file and folder names. I did this with a shell script that I hacked together from Stackoverflow answers, so it’s outside the scope of this article.
- Filtering for lines with valid email addresses: Many files in “Collection #1-5” are SQL dumps that contain much more than simple lines of email-password-combinations, some files also contained invalid email addresses or other garbage, so in the end I thought it would be best to only retain lines with valid email addresses.
- Split lines into email addresses and passwords (if they are delimited by a “;” or a “:”, which seemed to be the most common delimiters).
- Parse email addresses into their components such as “email_user”, “maindomain”, “subdomains”, “tld” (top-level domain such as “.ch”).
- A (simple) heuristic to remove lines with hashed passwords. This means that when I’m talking about 500 email addresses of the Swiss Armed Forces, these come with unhashed, plaintext passwords (which makes it, of course, more problematic).
Of course, all these steps involved a happy amount of Regex…
I packed everything into a batch routine which did the following:
- For each collection 1, 2, and 3-5 (they were all contained within their respective folders):
- Split up the files into batches of 2.5GB
- For each batch:
- Explicitly load the files from the HDD into memory
- Conduct the above preprocessing steps
- Save the batch as a so-called Parquet file onto the SSD
Collection 1, which is about 85GB large, for example, resulted in about 35 batches. I had a batch processing logic that would ignore failed batches and try them again in the next iteration. Sometimes, batches would fail for some magic reason and work in the next iteration . Also, often R would crash after a few hours (for no obvious reason, Spark almost never crashed and was still running then, and the memory was still half empty). After getting tired of restarting R manually, I developed an almost embarrassingly hacky cronjob that would look at the datetime of the last log entry and restart my R script automatically whenever that datetime was too long ago (e.g. a couple of hours).

Some explanations regarding the above steps:
- Point 1.1: 2.5GB of data can quickly become 20GB+ in memory and I had to make sure all the data fitted into memory during preprocessing. This is something I yet don’t fully understand but it has to do with the Java data structures Spark uses. So I found 2.5GB to be a good value, also because the probability of a batch failing decreased with its size.
- Point 1.2.3: Parquet files are special columnar data structures with a high compression rate. So a 2.5GB batch approximately resulted in a 1.5GB Parquet file. Also, reading from these files in the later steps turned out to be way faster than reading from CSV files, for example.
Why was I confident that this preprocessing logic worked (in a sufficiently accurate fashion)? For collection 1, for example, I saw that, after preprocessing, approximately 770 million unique email addresses remained. This is similar to the figure Troy Hunt reported initially. Besides that, I had other plausibility checks in place.
Filtering for the Swiss TLD
The result of this first major step were about 60GB of Parquet files for collection 1, 300GB for collection 2, 150GB for collection 3-5 and 50GB of data that I had to preprocess in a second iteration (split up into batches of 1GB to pinpoint some hard-to-debug errors). So that’s already a reduction to almost half of the initial size. I had to move this data back to the HDD to free up space on the SSD.
The next major step involved:
- For each mega-batch (collection 1, 2, …) on the HDD
- Filter for TLD == “.ch”
- Save the results as Parquet files on the SSD
Now you see what I was referring to when I talked about the 300GB+ job at the beginning of this post. The whole filtering and saving as Parquet file took about an hour for this huge amount of data. Spark handled the copying from HDD to memory and from there to SSD autonomously and without flaws, i.e. I never encountered an OOM error or the like. What’s important is that the main disk where Spark’s “tmp”-folder resides has enough space left. In my case, this was the SSD. Spark “spills” temporary data from memory to disk and this needs sufficient space – apart from the space that is needed for storing the resulting Parquet files. Make sure to use an SSD for disk spills because the whole process is so much slower on a HDD.
The analysis
In the last major step, I ended up with four Parquet files of a total size of around 1GB (31 million lines of Swiss addresses) and could process everything in-memory. This involved, among others:
- Counting distinct addresses (shown in the concrete example above)
- Searching for domains used by authorities and the like, and grouping by them
- Computing average and median password length and other stats
- Searching for keywords in email users (names of PEPs, for example)
- …
As shown above, this was very fast, so the second job above took about 5 seconds.

Summary and some learnings
While there are probably other, more efficient ways of doing this, I think that Spark and sparklyr really helped me to solve a “big data” problem that I wouldn’t have known how to deal with otherwise.
All in all, I spent a couple of weeks on this process, but once it ran in a robust fashion and I finally had dealt with all sources of errors, the preprocessing step took a couple of days to run, the filtering step a couple of hours and the analysis step a couple of minutes.
Without much further ado, I’d like to summarize some learnings and give some advice:
- Reduce data volume as early as possible. Filter out unnecessary stuff early on, save intermediate results in compressed form, etc. Sometimes this is not possible, and in my case, I’m still asking myself whether preprocessing couldn’t have been implemented more time-efficient.
- Invest enough time in a robust pipeline early on. By “robust” I mean a script that catches all sorts of errors and is guaranteed to not crash – a script that you don’t have to fondle and care for all the time. Try-catch is your friend. If you encounter some irreproducible fatal crashes on the R side, try to find a way to detect this and restart the process automatically. Also, once you have developed a good pipeline, you might not need RStudio anymore. It might be less error-prone to run scripts in the background compared to RStudio.
- Log to a file. I used the simple logger package to write all output to different files. This also allowed the hacky restarting routine, and made debugging easier (the R console becomes unavailable in RStudio once you encounter a joyful “R session terminated unexpectedly”).
- Try to understand the big data technology you’re using. In my case, I had the luck to recently complete a professional Spark training as part of a college “Data Science” diploma, but I still had to read up and understand stuff such as Spark tuning and optimization.
- Be willing to kill your darlings, e.g. use another language than R. During the whole process, I always asked myself whether it wouldn’t have been better to use PySpark from the very beginning. sparklyr works well and all, and I’m very familiar with R, but sometimes I had the feeling that PySpark has a better documentation and is “closer” to Spark than R is. Of course, Scala and Java would be even closer. Also, I was disappointed that R sometimes crashed for opaque reasons. However, in the end, I stayed true to my darling.
- Consider using the cloud. I didn’t do this for the outlined reason and because I have a nice workstation at hand, but it is a valid option. The cool thing about sparklyr is that you can easily switch to a cloud cluster: your R code doesn’t have to change at all. However, from my experience, working with the cloud requires some serious knowledge about the services available and the pricing models, for example. There are so many different options available and just choosing a good one for a certain problem might require quite some time as well.
All in all, I hope this will help data journalists and scientists alike when working with large volumes of data in the future, especially when processing data locally.
For comments, suggestions, questions, criticism and the like: Please give me a shout in the comment section or on Twitter.
